Posts
This week’s article is from the game-changer FLOT4 trial. I ll replicate a simple but very informative figure.
Selected article:
Title: Histopathological regression after neoadjuvant docetaxel, oxaliplatin, fluorouracil, and leucovorin versus epirubicin, cisplatin, and fluorouracil or capecitabine in patients with resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4-AIO): results from the phase 2 part of a multicentre, open-label, randomised phase 2/3 trial
Journal: Lancet Oncology
Authors: Al-Batran SE, Hofheinz RD, Pauligk C et al.
Year: 2016
PMID: 27776843
DOI: 10.1016/S1470-2045(16)30531-9
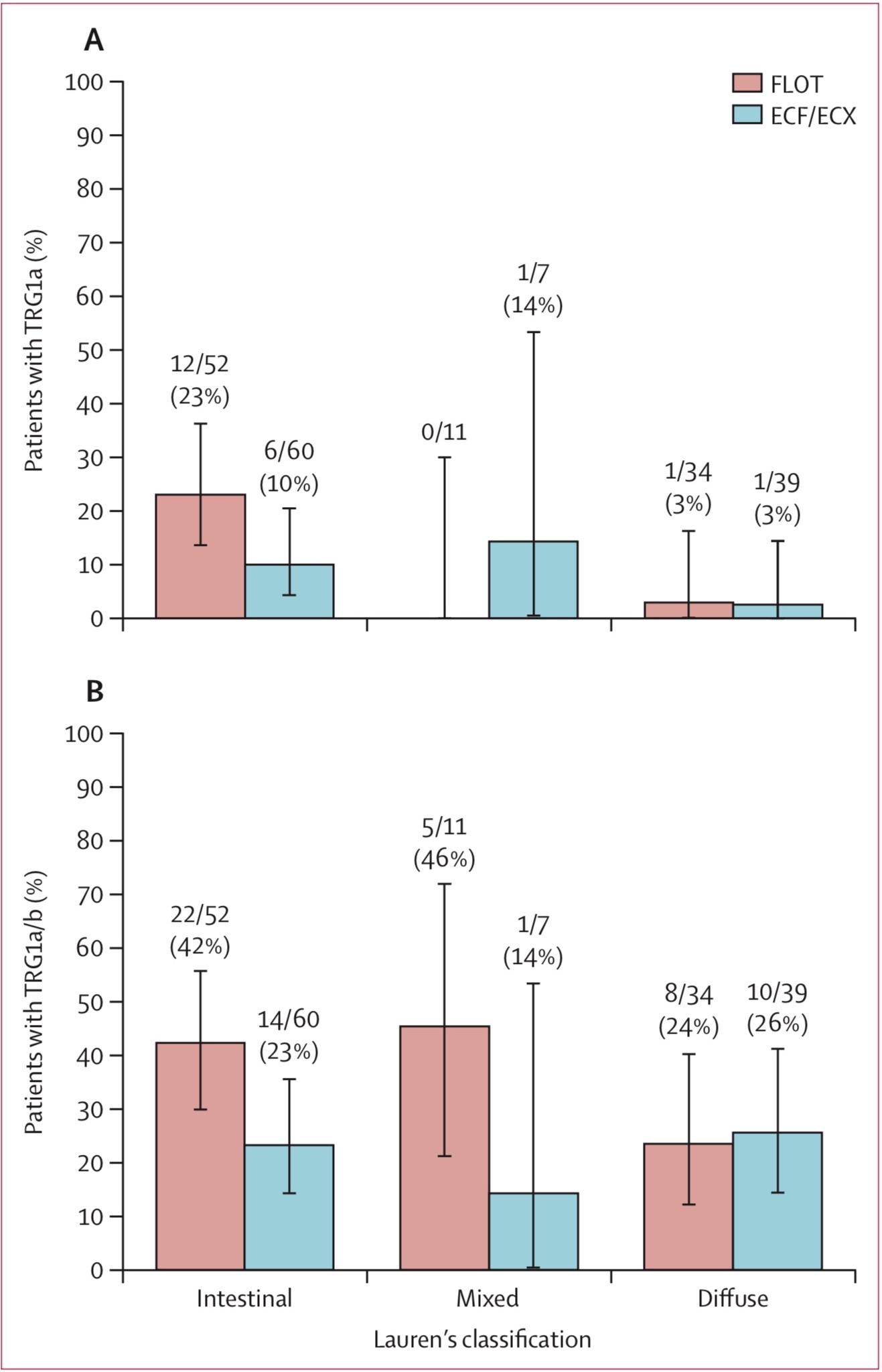
The original figure
Import libraries
library(tidyverse)
library(scales)
library(fabricatr) # to fabricate fake data
library(glue) # to prepare label text
library(broom) # to tidy prop.test()
library(lemon) # for facet_rep_wrap() function
theme_set(theme_light(base_family = "IBM Plex Sans"))
Prepare fabricated data
# FLOT
set.seed(2022)
FLOT_intestinal <- fabricate(
N = 52,
main_group = "FLOT",
lauren = "intestinal",
response = c(rep("1a", 12), rep("1b", 10), rep("others", N-22)))
FLOT_mixed <- fabricate(
N = 11,
main_group = "FLOT",
lauren = "mixed",
response = c(rep("1a", 0), rep("1b", 5), rep("others", N-5)))
FLOT_diffuse <- fabricate(
N = 34,
main_group = "FLOT",
lauren = "diffuse",
response = c(rep("1a", 1), rep("1b", 7), rep("others", N-8)))
# ECF/ECX
set.seed(2022)
ECF_intestinal <- fabricate(
N = 60,
main_group = "ECF/ECX",
lauren = "intestinal",
response = c(rep("1a", 6), rep("1b", 8), rep("others", N-14)))
ECF_mixed <- fabricate(
N = 7,
main_group = "ECF/ECX",
lauren = "mixed",
response = c(rep("1a", 1), rep("1b", 0), rep("others", N-1)))
ECF_diffuse <- fabricate(
N = 39,
main_group = "ECF/ECX",
lauren = "diffuse",
response = c(rep("1a", 1), rep("1b", 9), rep("others", N-10)))
# Combine datasets
combined_dataset <- bind_rows(
FLOT_intestinal, FLOT_mixed, FLOT_diffuse,
ECF_intestinal, ECF_mixed, ECF_diffuse,
) %>%
mutate(ID = paste0("p_" ,row_number()), .before = "main_group") %>%
as_tibble()
A part of fake dataset
set.seed(2022)
combined_dataset %>%
sample_n(10)
## # A tibble: 10 × 4
## ID main_group lauren response
## <chr> <chr> <chr> <chr>
## 1 p_179 ECF/ECX diffuse others
## 2 p_55 FLOT mixed 1b
## 3 p_75 FLOT diffuse others
## 4 p_196 ECF/ECX diffuse others
## 5 p_6 FLOT intestinal 1a
## 6 p_191 ECF/ECX diffuse others
## 7 p_123 ECF/ECX intestinal others
## 8 p_14 FLOT intestinal 1b
## 9 p_7 FLOT intestinal 1a
## 10 p_93 FLOT diffuse others
Possible strategy:
This time, I ll go with faceted figure.
I prefer using geom_col() instead of geom_bar(). Therefore, I ll do some calculations for percentage and CIs.
tidy_data <- combined_dataset %>%
group_by(main_group, lauren) %>%
summarise(n = n(),
n_1a = sum(response == "1a"),
n_1a1b = sum(response == "1a" | response == "1b")) %>%
ungroup() %>%
pivot_longer(cols = c("n_1a", "n_1a1b"),
names_to = "facet_group",
values_to = "values") %>%
rowwise() %>%
mutate(prop_table = list(tidy(prop.test(values, n, conf.level=0.95)))) %>%
unnest(prop_table) %>%
mutate(estimate = estimate * 100,
conf.low = conf.low * 100,
conf.high = conf.high * 100) %>% # mutate_at() or mutate(across()) is possible.
select(main_group:estimate, conf.low, conf.high) %>% # just to simplify the dataset
mutate_at(c("main_group", "lauren", "facet_group"), factor) %>%
mutate(main_group = fct_relevel(main_group, c("FLOT")),
lauren = fct_relevel(lauren, c("intestinal", "mixed", "diffuse"))) %>%
mutate(label_text = if_else(values == 0, glue("{values}/{n}"), glue("{values}/{n} \n({round(estimate)}%)")))
R codes for the figure
final_replica <- tidy_data %>%
ggplot(aes(lauren, estimate, fill = main_group)) +
geom_col(position = "dodge", colour="black", width = .7) +
geom_errorbar(aes(ymax = conf.high, ymin = conf.low), position = position_dodge(width = .7), width = .15) +
facet_rep_wrap(~ facet_group, ncol = 1, strip.position = "left", labeller = as_labeller(c(n_1a = "Patients with TRG1a (%)", n_1a1b = "Patients with TRG1a/b (%)"))) +
scale_y_continuous(limits = c(-2, 100), breaks = seq(0,100,10), expand = c(0, 0)) + # -2 was used to add outside ticks
scale_x_discrete(labels = c("Intestinal", "Mixed", "Diffuse")) +
scale_fill_manual(values = c("#D8A09A", "#9ED0D9")) +
geom_text(aes(y = conf.high + 8, label = label_text), position = position_dodge(width = .7), size = 4, family = "IBM Plex Sans") +
geom_hline(yintercept = 0, color = "black", size = .5) +
labs(y = "",
x = "Lauren's classification",
fill = "") +
theme(plot.margin = unit(c(.5, .5, .5, 0), "cm"),
plot.background = element_rect(colour = "#BC687F", fill = NA, size = .5),
strip.placement = "outside",
strip.background = element_blank(),
strip.text = element_text(color = "black", size = 11),
panel.grid = element_blank(),
panel.border = element_blank(),
panel.spacing = unit(1, "cm"),
axis.line = element_line(color = "black"),
axis.line.x = element_blank(),
axis.ticks.y = element_line(color = "black", size = .5),
axis.ticks.length.y = unit(.2, "cm"),
axis.text.y = element_text(size = 11, color = "black"),
axis.text.x = element_text(size = 11, color = "black"),
axis.title.x = element_text(vjust = -1.5),
legend.position = c(.85, .95),
legend.text = element_text(size = 12),
legend.key.height= unit(.4, "cm"),
legend.spacing.y = unit(.25, "cm"),
axis.ticks.x = element_blank()) +
guides(fill = guide_legend(byrow = TRUE)) +
geom_segment(aes(x = 1.5, xend = 1.5, y = 0, yend = -2)) +
geom_segment(aes(x = 2.5, xend = 2.5, y = 0, yend = -2)) +
geom_segment(aes(x = 3.5, xend = 3.5, y = 0, yend = -2))
### SAVE FIGURE
ggsave(final_replica,
file =file.path ("w7_replica.jpg"),
dpi = 300,
width = 6,
height = 9)
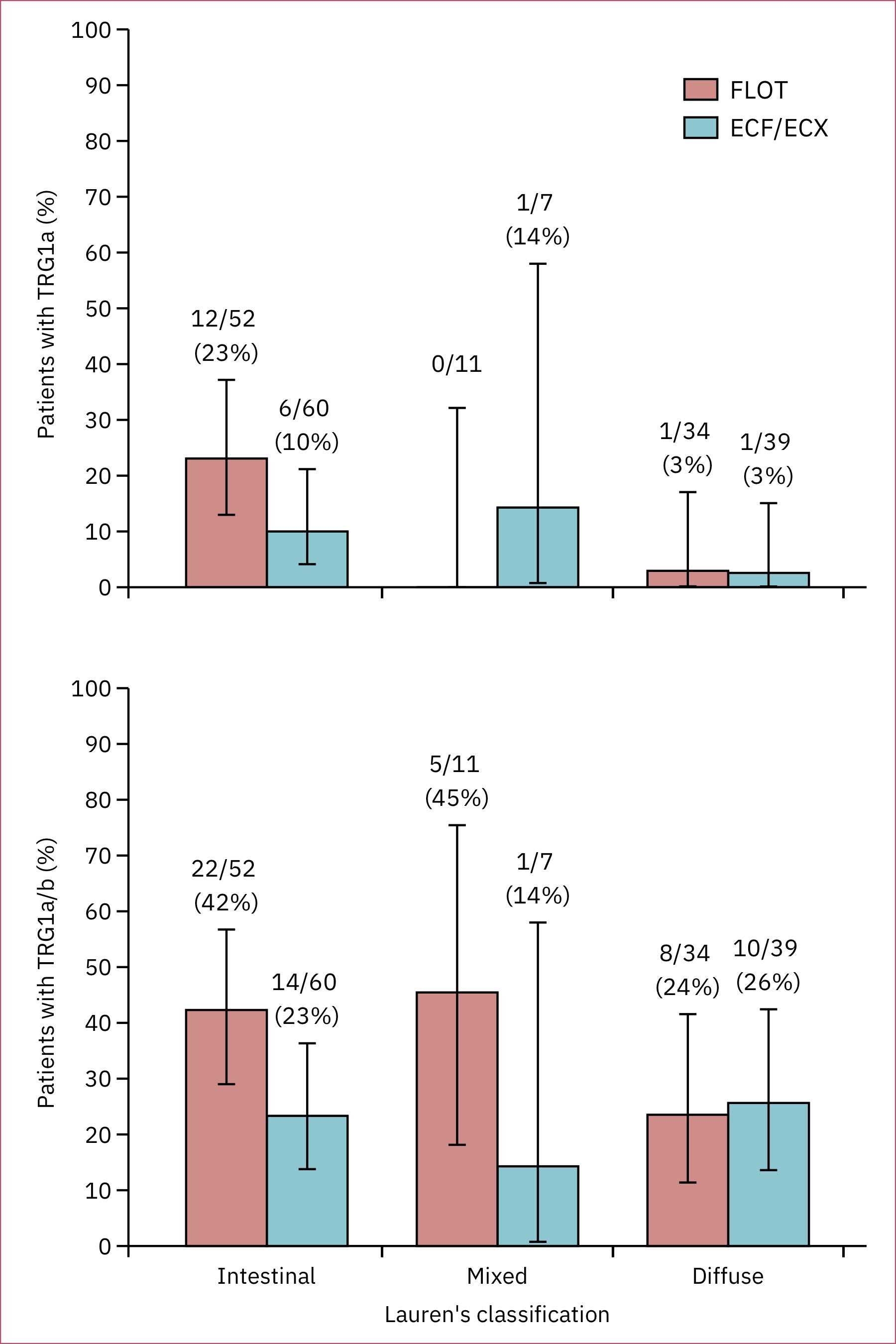
Final replica
Some personal comments:
Citation
Ali Guner (Apr 4, 2022) Week-7. Retrieved from https://datavizmed.com/blog/2022-04-04-week-7/
@misc{ 2022-week-7,
author = { Ali Guner },
title = { Week-7 },
url = { https://datavizmed.com/blog/2022-04-04-week-7/ },
year = { 2022 }
updated = { Apr 4, 2022 }
}